

set cartoon_highlight_color = grey50 # ray_trace_mode=1 makes a black outline around the secondary structural elements set ray_trace_mode = 1 # stick_radius -adjust thickness of atomic bonds set stick_radius = 0.2 # mesh_radius -to adjust thickness of electron density contours set mesh_radius = 0.02 # bg_color -set the background color bg_color white # load pdb file and give it an object name load csos3_18o_nobreak. #BEGIN PYMOL SCRIPT for Overall Fold -left half of stereo image # antialias =1 smooths jagged edges, 0 turns it off set antialias = 1 # Larger values of ambient make the image brighter set ambient = 0.3 # Larger values of direct eliminates shadows set direct = 1.0 set ribbon_radius = 0.2 # cartoon_highlight_color will give a separate color to edges of secondary structural elements. Otherwise, the secondary structure assignment made by pymol may be inaccurate.
#Pymol tutorial reaction download#
The script can be modified to produce the right half of the stereo image by changing the line "turn y, 3.5" to "turn y, -3.5"ģ) Download labeled figure in photoshop format.ġ) If you are modifying this script to illustrate a different set of coordinates, it is best to have the secondary structure assignments in the header. Depth is conveyed by use of fog, veiling less important structural features in the back of the enzyme.Ģ) Download Pymol script for left half of stereo image. Labels in the active site are given an “outer glow” to make them legible in a region of the figure that is dense in detail. Domain labels are colored to correspond to the domains they are labeling. SSEs are labeled directly on the individual SSEs. The orientation was chosen to feature the location of the active site (outlined by side chains and zinc ion), and to show the two-fold symmetry relationship between the active domain (yellow) and homologous but defunct domain (red).
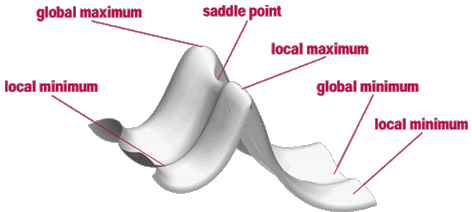
The three domains that compose this enzyme are distinguished by coloring each domain separately (blue, yellow, red). Illustration of the overall fold of carbonic anhydrase, CsoS3, from Halothiobacillus neapolitanus.(PDB ID 2G13). Special thanks to Steve Bottomley (Curtin Univ) for pointing-out that the relative orientation between panels B and C of Fig S3 is not clearly described the revised figure and caption attempt to clarify this point.
#Pymol tutorial reaction update#


 0 kommentar(er)
0 kommentar(er)
